#include <csm.hh>
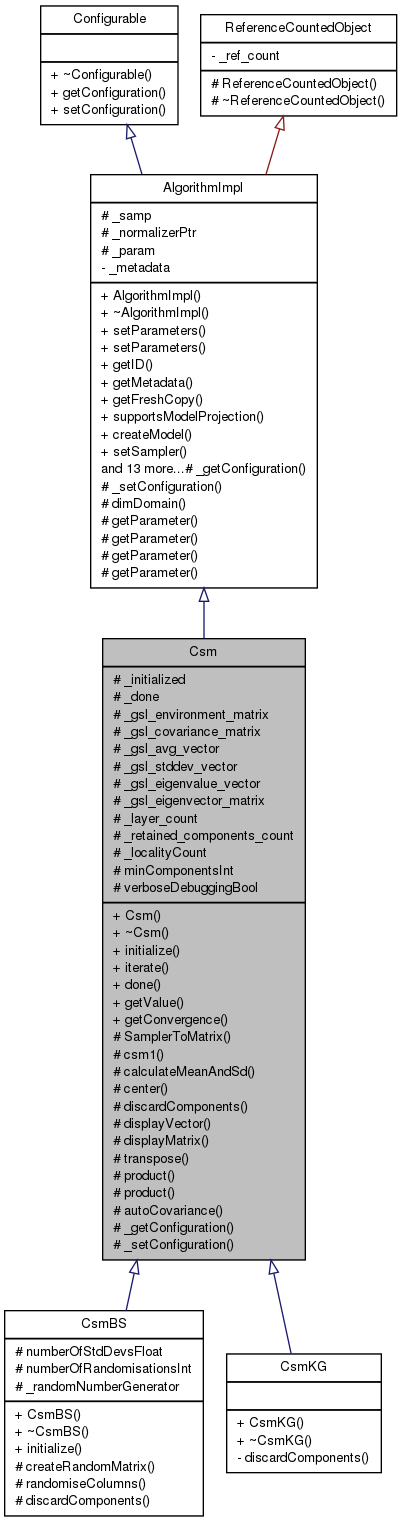
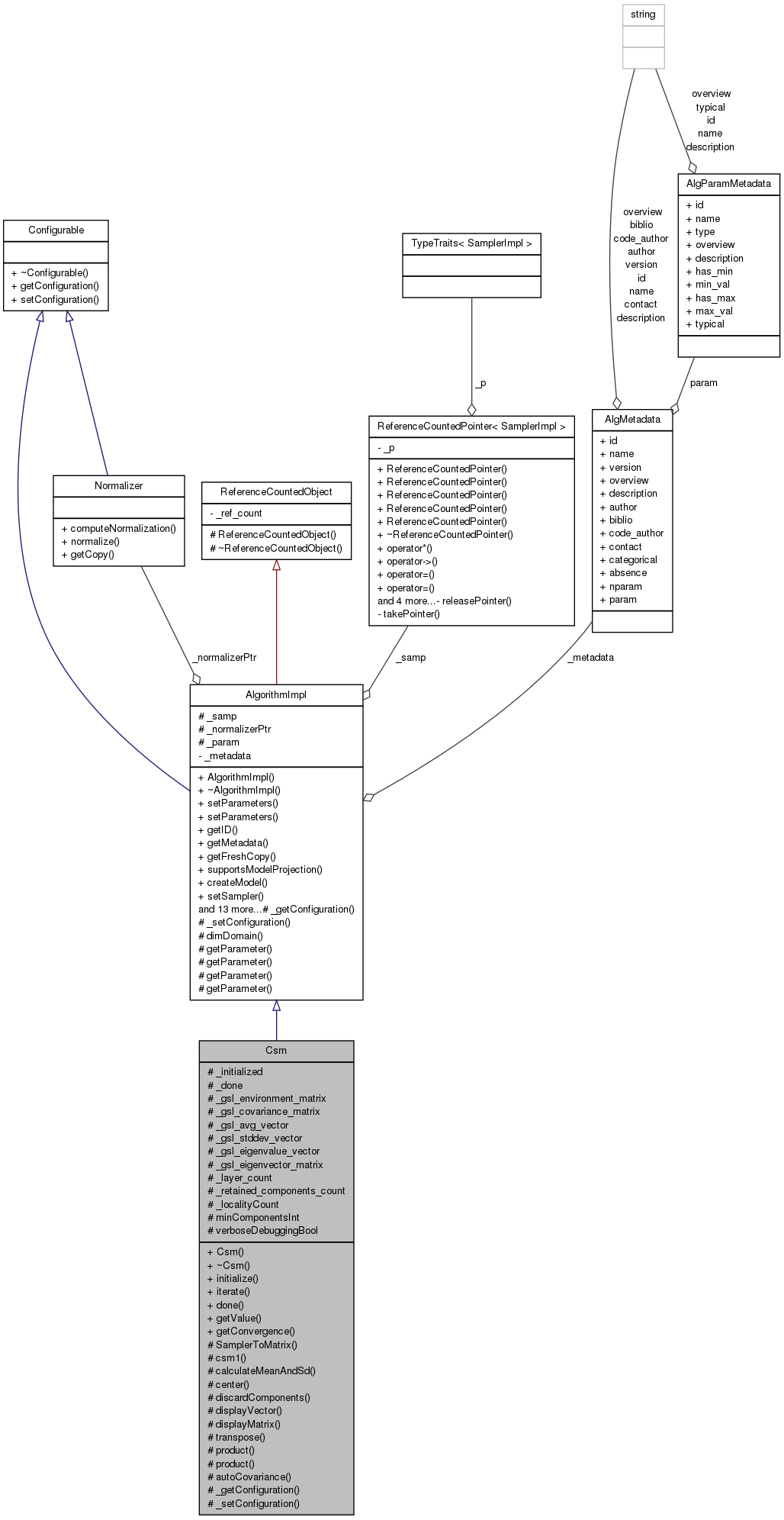
Public Member Functions | |
| Csm (AlgMetadata const *metadata) | |
| ~Csm () | |
| virtual int | initialize () |
| int | iterate () |
| int | done () const |
| Scalar | getValue (const Sample &x) const |
| int | getConvergence (Scalar *const val) const |
Protected Member Functions | |
| int | SamplerToMatrix () |
| bool | csm1 () |
| int | calculateMeanAndSd (gsl_matrix *theMatrix, gsl_vector *theMeanVector, gsl_vector *theStdDevVector) |
| int | center () |
| virtual int | discardComponents ()=0 |
| void | displayVector (const gsl_vector *v, const char *name, const bool roundFlag=true) const |
| void | displayMatrix (const gsl_matrix *m, const char *name, const bool roundFlag=true) const |
| gsl_matrix * | transpose (gsl_matrix *m) |
| double | product (gsl_vector *va, gsl_vector *vb) const |
| gsl_matrix * | product (gsl_matrix *a, gsl_matrix *b) const |
| gsl_matrix * | autoCovariance (gsl_matrix *m) |
| virtual void | _getConfiguration (ConfigurationPtr &config) const |
| virtual void | _setConfiguration (const ConstConfigurationPtr &config) |
Protected Attributes | |
| int | _initialized |
| int | _done |
| gsl_matrix * | _gsl_environment_matrix |
| gsl_matrix * | _gsl_covariance_matrix |
| gsl_vector * | _gsl_avg_vector |
| gsl_vector * | _gsl_stddev_vector |
| gsl_vector * | _gsl_eigenvalue_vector |
| gsl_matrix * | _gsl_eigenvector_matrix |
| int | _layer_count |
| int | _retained_components_count |
| int | _localityCount |
| int | minComponentsInt |
| bool | verboseDebuggingBool |
Detailed Description
Herewith follows a detailed explanation of the Climate Space Model (CSM). Note that the CSM model was developed by Dr Neil Caithness. This implementation of CSM was written by Tim Sutton and Renato De Giovanni.
//////////////////////////////////////////////////////
// Model Creation
//////////////////////////////////////////////////////
Inputs:
File contiaing xy point localties
List of gdal layers
----------------------------------
Look up values at each locality in each layer
|x|y|var1|var2 |var3 |.... |varN |
-------------------------------------------
1 | | | | | | | |
-------------------------------------------
2 | | | | | | | |
-------------------------------------------
3 | | | | | | | |
-------------------------------------------
4 | | | | | | | |
-------------------------------------------
5 | | | | | | | |
-------------------------------------------
6 | | | | | | | |
-------------------------------------------
7 | | | | | | | |
-------------------------------------------
8 | | | | | | | |
-------------------------------------------
etc.
Now remove any rows with nans in (GDAL NO_DATA)
Now remove any rows which are duplicates {optional step!]
After duplicates have been removed, lat and long columns can be removed.
Now we need to center and standardise the data (auto)
Before:
. . |
. .. |
. . . |
|
-----------------
. |.
. |
. . |
.. |
After:
|
|
.|.
..|..
-----------------
..|..
.|.
|
|
To do this:
Calculate the mean for every column (excluding lat/long)
Calculate the stddev for every column (excluding lat/long)
Subtract the column mean from every value in that column
Divide each restultant column value by the stddev for that column
Make sure you remember the column stddev and mean for each column for later use.
Now calculate the covariance matrix:
Pass the data matrix to a covariance function (e.g. in GSL?) - note the datamatrix
should not include the column stddev and mean values.
The resulting covariance matrix will have the same number of rows as columns i.e. it is square.
Note that the data in the covariance matrix no longer resembles the input point
lookup data!
-----------------------------------------
|var1|var2 |var3 |.... |varN |
-----------------------------------------
1 | | | | | |
-----------------------------------------
2 | | | | | |
-----------------------------------------
3 | | | | | |
-----------------------------------------
... | | | | | |
-----------------------------------------
N | | | | | |
-----------------------------------------
Now obtain the eigenvalues and eigenvector of the covariance matrix using GSL
The eigenvector will look something like this:
-------------------------------------------
| 1 | 2 | 3 |.... |component N |
-------------------------------------------
Var 1 | | | | | |
-------------------------------------------
Var 2 | | | | | |
-------------------------------------------
Var 3 | | | | | |
-------------------------------------------
..... | | | | | |
-------------------------------------------
Var N | | | | | |
-------------------------------------------
Each column represents one component, and each row represents one of the input variables transposed
order of original covariance matrix columns.
The cell values represent the loading / weight of that variable in that component.
The eigenvalues are the values through the diagonal of the output of the eigenvalues funtion. (prefixed with x above)
-------------------------------------------
| 1 | 2 | 3 |.... |component N |
-------------------------------------------
Var 1 | x5 | 0 | 0 | 0 | 0 |
-------------------------------------------
Var 2 | 0 | x8 | 0 | 0 | 0 |
-------------------------------------------
Var 3 | 0 | 0 | x1 | 0 | 0 |
-------------------------------------------
..... | 0 | 0 | 0 | x4 | 0 |
-------------------------------------------
Var N | 0 | 0 | 0 | 0 | xN |
-------------------------------------------
This is a separate vector to the one created by the eigenvector function.
The sum of the eigenvalues should be equal to the number of columns!
Next we arrange the column order of the eigenvector according to the descending values of the
eigenvalues.
-------------------------------------------
| 2 | 1 | 4 |.... |component N |
-------------------------------------------
Var 1 | x8 | | | | |
-------------------------------------------
Var 2 | | x5 | | | |
-------------------------------------------
Var 3 | | | x4 | | |
-------------------------------------------
..... | | | | x1 | |
-------------------------------------------
Var N | | | | | xN |
-------------------------------------------
The next step is to remove any column from the eigenvector
where the eigenvalue is less than 1 (in the kaiser-gutman method), or
to remove any column where the eigenvalue is less than a randomised
cutoff) broken stick method.
-------------------------------------
| 2 | 1 | 4 |component N |
-------------------------------------
Var 1 | | | | |
-------------------------------------
Var 2 | | | | |
-------------------------------------
Var 3 | | | | |
-------------------------------------
..... | | | | |
-------------------------------------
Var N | | | | |
-------------------------------------
That complete the CSM model definition
//////////////////////////////////////////////////////
// Model Projection:
//////////////////////////////////////////////////////
Inputs:
Data layers that will be used as the basis for the model projection (must match the dimensions and units of the input dataset).
The standard deviation for each of the layers as calculated in the model definition process.
The mean of each layer as calculated in the model definition process.
Now for each layer visit each cell, subtract the mean (xbar) and divide the result by the standard deviation.
This step is called 'auto'.
Note these must be the mean and standard deviation particular to that layer as calculated in the model definition process.
Next we create the scores.
This is carried out by performing matrix multiplication - multiplying the independent variable layers (produced by auto above) by the eigenvectors.
The output is one new 'layer' (actually a component) for each of the components kept during the model building process.
Layers after auto
+----------------+
|a | Layer 1
| + - - - - |---------+
| | b | | Layer 2
| | | .
| | | | .
+----------------+ | Layer n
| |
| |
+-------------------+
-------------------------------------
| 2 | 1 | 4 |component N |
-------------------------------------
Layer 1 | | | | |
-------------------------------------
Layer 2 | | | | |
-------------------------------------
Layer 3 | | | | |
-------------------------------------
..... | | | | |
-------------------------------------
Var N | | | | |
-------------------------------------
Constructor & Destructor Documentation
| Csm::Csm | ( | AlgMetadata const * | metadata | ) |
Constructor for Csm
Constructor for Csm
- Parameters:
-
Sampler is class that will fetch environment variable values at each occurrence / locality
Definition at line 57 of file csm.cpp.
References _initialized, and verboseDebuggingBool.
| Csm::~Csm | ( | ) |
This is the descructor for the Csm class
Definition at line 70 of file csm.cpp.
References _gsl_avg_vector, _gsl_covariance_matrix, _gsl_eigenvalue_vector, _gsl_eigenvector_matrix, _gsl_environment_matrix, _gsl_stddev_vector, and _initialized.
Member Function Documentation
| void Csm::_getConfiguration | ( | ConfigurationPtr & | config | ) | const [protected, virtual] |
Method to serialize a CSM model.
- Parameters:
-
config Pointer to the serializer object
Reimplemented from AlgorithmImpl.
Definition at line 723 of file csm.cpp.
References _done, _gsl_avg_vector, _gsl_eigenvalue_vector, _gsl_eigenvector_matrix, _gsl_stddev_vector, _layer_count, and _retained_components_count.
| void Csm::_setConfiguration | ( | const ConstConfigurationPtr & | config | ) | [protected, virtual] |
Method to deserialize a CSM model.
- Parameters:
-
config Pointer to the serializer object
Reimplemented from AlgorithmImpl.
Definition at line 770 of file csm.cpp.
References _done, _gsl_avg_vector, _gsl_eigenvalue_vector, _gsl_eigenvector_matrix, _gsl_stddev_vector, _layer_count, and _retained_components_count.
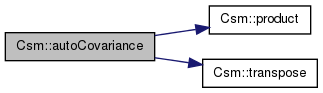
| gsl_matrix * Csm::autoCovariance | ( | gsl_matrix * | original_matrix | ) | [protected] |
This a utility function to calculate the auto covariance of a gsl matrix.
- Parameters:
-
m gsl_matrix Input matrix
- Returns:
- gsl_matrix Output matrix
This method tries to mimic the octave "cov" function when it receives only one parameter:
function c = cov (x)
if (rows (x) == 1) x = x'; endif
n = rows (x);
x = x - ones (n, 1) * sum (x) / n; c = conj (x' * x / (n - 1));
endfunction
Definition at line 578 of file csm.cpp.
References product(), and transpose().
Referenced by csm1(), and CsmBS::discardComponents().
| int Csm::calculateMeanAndSd | ( | gsl_matrix * | theMatrix, |
| gsl_vector * | theMeanVector, | ||
| gsl_vector * | theStdDevVector | ||
| ) | [protected] |
Calculate the mean and standard deviation of the environment variables at the occurence points.
- Note:
- The matrix, mean and stddev vectors MUST be pre-initialised!
- Parameters:
-
theMatrix - a gsl_matrix pointer from which mean and stddev will be obtained theMeanVector - a pointer to a gsl_vector in which the column means will be stored theStdDevVector - a pointer to a gsl_vector in which the column stddevs will be stored
- Returns:
- 0 on error
NOTE: the mean and stddev vectors MUST be pre-initialised!
Definition at line 175 of file csm.cpp.
References _layer_count.
Referenced by csm1(), and CsmBS::discardComponents().
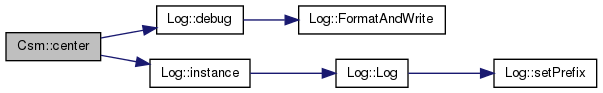
| int Csm::center | ( | ) | [protected] |
Center and standardise. Subtract the column mean from every value in each column Divide each resultant column value by the stddev for that column
- Note:
- This method must be called after calculateMeanAndSd
- Returns:
- 0 on error
Definition at line 217 of file csm.cpp.
References _gsl_avg_vector, _gsl_environment_matrix, _gsl_stddev_vector, _layer_count, _localityCount, Log::debug(), and Log::instance().
Referenced by csm1().
| bool Csm::csm1 | ( | ) | [protected] |
This is a wrapper to call several of the methods below to generate the initial model.
Csm1 is used to produce the model definition
Definition at line 648 of file csm.cpp.
References _gsl_avg_vector, _gsl_covariance_matrix, _gsl_eigenvalue_vector, _gsl_eigenvector_matrix, _gsl_environment_matrix, _gsl_stddev_vector, _layer_count, _retained_components_count, autoCovariance(), calculateMeanAndSd(), center(), Log::debug(), discardComponents(), Log::instance(), and Log::warn().
Referenced by initialize().
| virtual int Csm::discardComponents | ( | ) | [protected, pure virtual] |
Discard unwanted components. This is a pure virtual function - it must be implemented by the derived class. Currently two derived classes are expected to be implemented - one for kaiser-gutman cutoff and one for broken-stick cutoff.
- Note:
- This method must be called after center
- Returns:
- 0 on error
Implemented in CsmBS, and CsmKG.
Referenced by csm1().
| void Csm::displayMatrix | ( | const gsl_matrix * | m, |
| const char * | name, | ||
| const bool | roundFlag = true |
||
| ) | const [protected] |
This a utility function to display the content of a gsl matrix.
- Parameters:
-
m gsl_matrix Input matrix name char Matrix name / message roundFlag Whether to round numbers to 4 decimal places (default is true)
Definition at line 451 of file csm.cpp.
References verboseDebuggingBool.
Referenced by CsmBS::discardComponents(), and getValue().
| void Csm::displayVector | ( | const gsl_vector * | v, |
| const char * | name, | ||
| const bool | roundFlag = true |
||
| ) | const [protected] |
This a utility function to display the content of a gsl vector.
- Parameters:
-
v gsl_vector Input vector name char Vector name / message roundFlag Whether to round numbers to 4 decimal places (default is true)
Definition at line 413 of file csm.cpp.
References verboseDebuggingBool.
Referenced by CsmBS::discardComponents(), and getValue().
| int Csm::done | ( | ) | const [virtual] |
Use this method to find out if the model has completed (e.g. convergence point has been met.
- Note:
- This method is inherited from the Algorithm class
- Returns:
- Implementation specific but usually 1 for completion.
Reimplemented from AlgorithmImpl.
Definition at line 268 of file csm.cpp.
References _done.
| int Csm::getConvergence | ( | Scalar *const | val | ) | const [virtual] |
Returns a value that represents the convergence of the algorithm expressed as a number between 0 and 1 where 0 represents model completion.
- Returns:
- Parameters:
-
val
Returns a value that represents the convergence of the algorithm expressed as a number between 0 and 1 where 0 represents model completion.
- Returns:
- Parameters:
-
Scalar *val
Reimplemented from AlgorithmImpl.
| Scalar Csm::getValue | ( | const Sample & | x | ) | const [virtual] |
This method is used when projecting the model.
- Note:
- This method is inherited from the Algorithm class
- Returns:
- Scalar of the probablitiy of occurence
- Parameters:
-
x Pointer to a vector of openModeller Scalar type (currently double). The vector should contain values looked up on the environmental variable layers into which the mode is being projected.
This method is used when projecting the model.
- Note:
- This method is inherited from the Algorithm class
- Returns:
- Scalar of the probablitiy of occurence must be between 0 and 1
- Parameters:
-
Scalar *x a pointer to a vector of openModeller Scalar type (currently double). The vector should contain values looked up on the environmental variable layers into which the mode is being projected.
Implements AlgorithmImpl.
Definition at line 283 of file csm.cpp.
References _gsl_avg_vector, _gsl_eigenvalue_vector, _gsl_eigenvector_matrix, _gsl_stddev_vector, _layer_count, displayMatrix(), displayVector(), Log::instance(), product(), verboseDebuggingBool, and Log::warn().
| int Csm::initialize | ( | ) | [virtual] |
Initialise the model specifying a threshold / cutoff point. This is optional (model dependent).
- Note:
- This method is inherited from the Algorithm class
- Returns:
- 0 on error
Initialise the model specifying a threshold / cutoff point. Any model definition building stuff is done here. This is optional (model dependent).
- Note:
- This method is inherited from the Algorithm class
- Parameters:
-
@return 0 on error
Implements AlgorithmImpl.
Reimplemented in CsmBS.
Definition at line 98 of file csm.cpp.
References _initialized, _layer_count, _localityCount, AlgorithmImpl::_samp, csm1(), Log::debug(), Log::instance(), SamplerToMatrix(), and Log::warn().
| int Csm::iterate | ( | ) | [virtual] |
Start model execution (build the model).
- Note:
- This method is inherited from the Algorithm class
- Returns:
- 0 on error
Reimplemented from AlgorithmImpl.
Definition at line 255 of file csm.cpp.
References _done.
| double Csm::product | ( | gsl_vector * | va, |
| gsl_vector * | vb | ||
| ) | const [protected] |
This a utility function to calculate the internal product of two gsl vectors.
- Parameters:
-
va gsl_vector Input vector a vb gsl_vector Input vector b
- Returns:
- double Result
Definition at line 510 of file csm.cpp.
Referenced by autoCovariance(), getValue(), and product().
| gsl_matrix * Csm::product | ( | gsl_matrix * | a, |
| gsl_matrix * | b | ||
| ) | const [protected] |
| int Csm::SamplerToMatrix | ( | ) | [protected] |
This is a utility function to convert a Sampler to a gsl_matrix.
- Returns:
- 0 on error
This is a utility function to convert the _sampl Sampler to a gsl_matrix.
- Returns:
- 0 on error
Definition at line 142 of file csm.cpp.
References _gsl_environment_matrix, _layer_count, _localityCount, AlgorithmImpl::_samp, Log::debug(), and Log::instance().
Referenced by initialize().
| gsl_matrix * Csm::transpose | ( | gsl_matrix * | m | ) | [protected] |
This a utility function to calculate a transposed gsl matrix.
- Parameters:
-
m gsl_matrix Input matrix
- Returns:
- gsl_matrix Transposed matrix
Definition at line 493 of file csm.cpp.
Referenced by autoCovariance().
Member Data Documentation
int Csm::_done [protected] |
This member variable is used to indicate whether the model building process has completed yet.
Definition at line 426 of file csm.hh.
Referenced by _getConfiguration(), _setConfiguration(), done(), and iterate().
gsl_vector* Csm::_gsl_avg_vector [protected] |
This is a pointer to a gsl vector that will hold the mean of each environmental variable column
Definition at line 436 of file csm.hh.
Referenced by _getConfiguration(), _setConfiguration(), center(), csm1(), getValue(), and ~Csm().
gsl_matrix* Csm::_gsl_covariance_matrix [protected] |
gsl_vector* Csm::_gsl_eigenvalue_vector [protected] |
This is a pointer to a gsl vector that will hold the eigen values
Definition at line 440 of file csm.hh.
Referenced by _getConfiguration(), _setConfiguration(), csm1(), CsmKG::discardComponents(), CsmBS::discardComponents(), getValue(), and ~Csm().
gsl_matrix* Csm::_gsl_eigenvector_matrix [protected] |
This is a pointer to a gsl matrix that will hold the eigen vectors
Definition at line 442 of file csm.hh.
Referenced by _getConfiguration(), _setConfiguration(), csm1(), CsmKG::discardComponents(), CsmBS::discardComponents(), getValue(), and ~Csm().
gsl_matrix* Csm::_gsl_environment_matrix [protected] |
This is a pointer to a gsl matrix containing the 'looked up' environmental variables at each locality. It is converted to a gsl matrix from the oM Sampler.samples primitive structure.
Definition at line 430 of file csm.hh.
Referenced by center(), csm1(), CsmBS::discardComponents(), SamplerToMatrix(), and ~Csm().
gsl_vector* Csm::_gsl_stddev_vector [protected] |
This is a pointer to a gsl vector that will hold the stddev of each environmental variable column
Definition at line 438 of file csm.hh.
Referenced by _getConfiguration(), _setConfiguration(), center(), csm1(), getValue(), and ~Csm().
int Csm::_initialized [protected] |
This is a flag to indicate that the algorithm was initialized.
Definition at line 423 of file csm.hh.
Referenced by Csm(), CsmBS::CsmBS(), CsmKG::CsmKG(), initialize(), and ~Csm().
int Csm::_layer_count [protected] |
Dimension of environmental space.
Definition at line 444 of file csm.hh.
Referenced by _getConfiguration(), _setConfiguration(), calculateMeanAndSd(), center(), csm1(), CsmKG::discardComponents(), CsmBS::discardComponents(), getValue(), initialize(), and SamplerToMatrix().
int Csm::_localityCount [protected] |
the number of localities used to construct the model
Definition at line 448 of file csm.hh.
Referenced by center(), initialize(), and SamplerToMatrix().
int Csm::_retained_components_count [protected] |
Number of components that are actually kept after Keiser-Gutman test
Definition at line 446 of file csm.hh.
Referenced by _getConfiguration(), _setConfiguration(), csm1(), CsmKG::discardComponents(), and CsmBS::discardComponents().
int Csm::minComponentsInt [protected] |
Minumum number of components required for a valid model
Definition at line 451 of file csm.hh.
Referenced by CsmBS::discardComponents(), and CsmBS::initialize().
bool Csm::verboseDebuggingBool [protected] |
Whether verbose debugging is enabled
Definition at line 453 of file csm.hh.
Referenced by Csm(), displayMatrix(), displayVector(), getValue(), and CsmBS::initialize().
The documentation for this class was generated from the following files:

